Go Beyond the Reference Genome.
Virus-Mapper unlocks the full story of your viral data. Instantly classify variants against a pangenome to discover novel mutations and track lineage-defining haplotypes with unprecedented clarity.
Get Early Access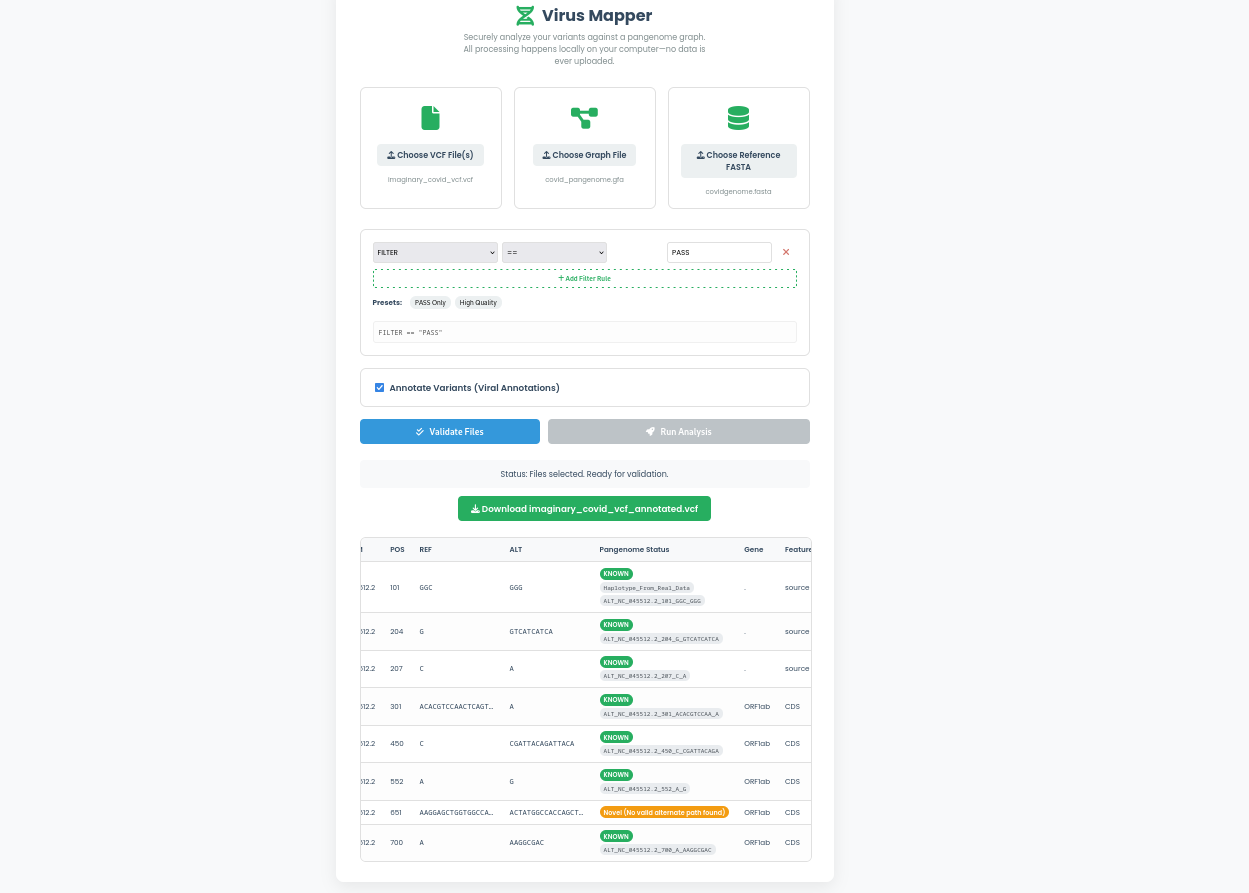
From Raw Data to Actionable Insight
Upload Your Data
Provide your VCF file, a pangenome graph (GFA/ODGI), and its corresponding reference FASTA. Our pre-flight checks ensure data compatibility.
Run Analysis
Our high-performance Rust engine interrogates every variant against the graph, leveraging the power of ODGI for maximum speed and efficiency.
Get Annotated Results
Download an enriched VCF file with clear "Known" vs. "Novel" classifications, haplotype information, and optional viral gene annotations.
Ready to Join the Program? 🚀
Get started with our secure, local-first Docker installation and see what you've been missing in your variant data.
View Installation Guide